It's been a long time, since I wrote a post here. Finally, final year of my course and so very busy :) with books! And now, got time to share with you.
As you know already (don't know?, then get to know :P ), I use to have favorite subjects in each semester, third semester it was microbiology, then in fourth semester, it was cell biology, fifth was molecular biology, sixth was genetic engineering and now i'm in seventh semester, but, you know what, I don't have a favorite subject!!! Don't worry that I lost interest, instead, I got 3 favorite subjects - Immunology, Animal biotechnology and Plant Biotechnology!
It's awesome this time to have more than one favorite subject and here I'm gonna tell you something about, cre-lox system which is most widely used for making modifications (mostly deletions). These modifications can be done at a specific tissue alone by using a tissue specific promoter i.e., you can selectively knock out a particular gene in particular cells (for eg: hepatocytes) alone.
Cre recombinase is a protein which could bind with specific sequences called loxP sequences. To explain it in a simple way, the cre recombinase binds at loxP sites on either side of your gene (which is to be deleted) and makes a cut! This leads to removal of the gene and further the break is repaired using the DNA ligase of host system.
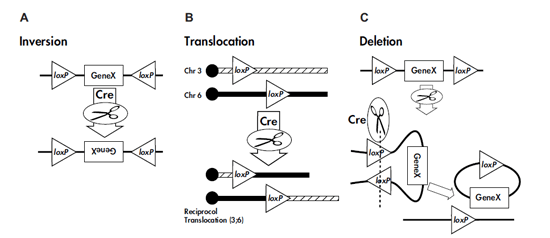
A single loxP site will have 13bp on either side of middle 8bp. The cre recombinase protein binds with this 13bp sequences forming dimer i.e., one loxP site contributes for a dimer (containing 2 cre recombinase units). Similarly, loxP on the other side of the target gene will contribute for a dimer formation. Then these two dimers will form tetramer (by folding the DNA strand as shown in figure. Then, cutting at the tetramer site happens leading to either translocation, inversion or deletion depending on the direction of the loxP site (i.e whether it is direct or invert repeat on either side of the target gene or located on different chromosomes)
13bp 8bp 13bp
ATAACTTCGTATA -NNNTANNN-TATACGAAGTTAT
This insertion, deletion and translocation could be done specifically at a particulat type of cell by using specific promoters. For example: To delete a particular gene in kidney cells alone, one could use kidney specific promoter for controlling the production of cre recombinase, so that cre recombinase will be expressed only in kidney cells. The molecular mechanisms of this deletions and recombinations invloves holliday junction formation. But, to be frank, i'm not clear or good at this holliday junction.
I''m trying to understand the molecular mechanism behind this, and, once I'm clear with it, I'll share it with you.
Understood the cre-loxp system? Got any doubt? anything wrong in my explanation? Kindly comment and I would try to reply or correct as soon as possible.
As you know already (don't know?, then get to know :P ), I use to have favorite subjects in each semester, third semester it was microbiology, then in fourth semester, it was cell biology, fifth was molecular biology, sixth was genetic engineering and now i'm in seventh semester, but, you know what, I don't have a favorite subject!!! Don't worry that I lost interest, instead, I got 3 favorite subjects - Immunology, Animal biotechnology and Plant Biotechnology!
It's awesome this time to have more than one favorite subject and here I'm gonna tell you something about, cre-lox system which is most widely used for making modifications (mostly deletions). These modifications can be done at a specific tissue alone by using a tissue specific promoter i.e., you can selectively knock out a particular gene in particular cells (for eg: hepatocytes) alone.
Cre recombinase is a protein which could bind with specific sequences called loxP sequences. To explain it in a simple way, the cre recombinase binds at loxP sites on either side of your gene (which is to be deleted) and makes a cut! This leads to removal of the gene and further the break is repaired using the DNA ligase of host system.
A single loxP site will have 13bp on either side of middle 8bp. The cre recombinase protein binds with this 13bp sequences forming dimer i.e., one loxP site contributes for a dimer (containing 2 cre recombinase units). Similarly, loxP on the other side of the target gene will contribute for a dimer formation. Then these two dimers will form tetramer (by folding the DNA strand as shown in figure. Then, cutting at the tetramer site happens leading to either translocation, inversion or deletion depending on the direction of the loxP site (i.e whether it is direct or invert repeat on either side of the target gene or located on different chromosomes)
13bp 8bp 13bp
ATAACTTCGTATA -NNNTANNN-TATACGAAGTTAT
This insertion, deletion and translocation could be done specifically at a particulat type of cell by using specific promoters. For example: To delete a particular gene in kidney cells alone, one could use kidney specific promoter for controlling the production of cre recombinase, so that cre recombinase will be expressed only in kidney cells. The molecular mechanisms of this deletions and recombinations invloves holliday junction formation. But, to be frank, i'm not clear or good at this holliday junction.
I''m trying to understand the molecular mechanism behind this, and, once I'm clear with it, I'll share it with you.
Understood the cre-loxp system? Got any doubt? anything wrong in my explanation? Kindly comment and I would try to reply or correct as soon as possible.
Lovely Your Post..............
ReplyDeleteCRE Recombinase- GenTarget Inc - CRE Recombinase, from bacteriophage P1, catalyzes recombination between 34 base-pair target sequences, called lox sites. Purified CRE enzyme can join individual plasmids containing lox sites.
Nicely explained. I am also writing a blogs on life sciences topics. Kindly go through them and give me some suggestions regarding my blogs.
ReplyDeletehttp://netweget.blogspot.in/